Protein-Protein interactions:
Using explicit water molecular dynamics simulations and
free energy estimates we try to understand the mechanisms
of protein recognition.
Our efforts have recently unveiled
a new view on this subject, emphasizing the role of desolvation as a
dominant force in recognition. We have also found
that nanoscale receptor-ligand interactions stemming from
electrostatic and solvation effects tunes the association
time scales over a range spanning as much as 5 orders of
magnitude in complexes where long electrostatics provide the specificty for recognition. Desolvation mediated complexes only span 3 order magnitude.
Simulations have explicitly revealed that
recognition arises from an optimized molecular key already
encoded in the protein's primary structure. Similarly, well
defined non-covalent, and non-specific, bonds are responsible for
locking the encounter complex in place.
See Anchor residues in protein-protein interactions,
PNAS 2004
[1]|[2]|[3]
|

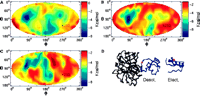
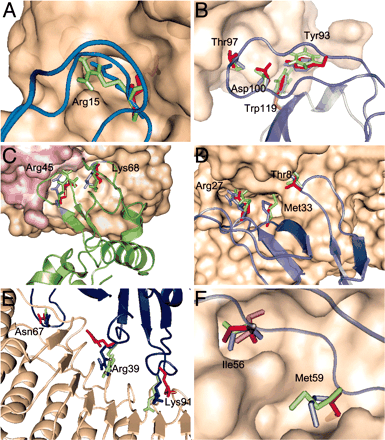 Anchor Residues in Protein Recognition, PNAS 2004
Anchor Residues in Protein Recognition, PNAS 2004
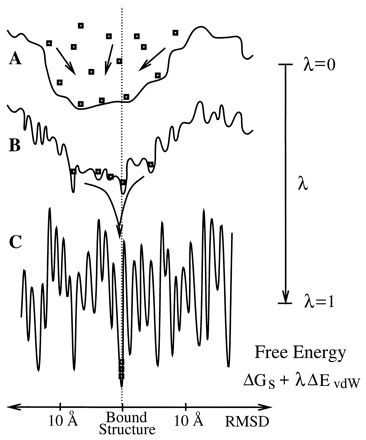 SmoothDock, PNAS 2001
SmoothDock, PNAS 2001